
 |
| Home | HLAsupB | Promiscuous Presentation | PromPDD | Publication | Tutorial | Download |
 |
|
|
Tutorials
|
| I. Architecture of HLAsupE |
| II. Definition of HLA supertypes |
| III. Usage of HLAsupE |
| i. Search for HLA supertype-specific epitopes or peptides |
| ii. Map HLA supertype-specific epitopes or peptides on an input protein sequence |
| iii. Search for mutated analogues of an input peptide |
| iv. Search for the promiscuous peptides presented by HLA molecules across supertypes |
| IV. Notes |
|
|
|
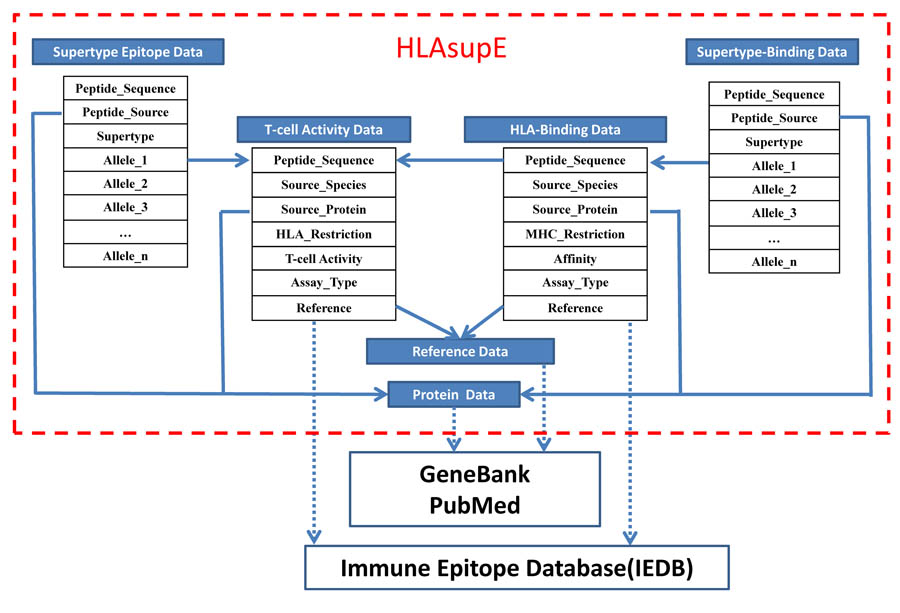
Architecture of HLAsupE
|
 |
|
|
|
Definition of HLA supertypes in HLAsupE
|
The HLA class I supertypes for the HLA-A and B loci used here were defined by Sidney et al (2008), and the supertypes used for the HLA-C loci were based on the classification presented by Doytchinova (2004). The HLA class II supertypes used were consistent with Doytchinova’s definition (2005). Due to the relatively low number of HLA-DPA and DPB alleles with available peptide data, all available HLA-DPA and DPB alleles were classified into one DP supertype. The available supertypes and corresponding alleles can be found on the download page of HLAsupE. |
|
|
|
Usage of HLAsupE
|
|
|
|
Search for HLA suppertype specific epitopes or peptidese
|
|
“peptide sequence”: amino acid sequence coding with uppercase letters. example : "FLPSDFFPSV"
|
|
|
|
Map HLA supertype-specific epitopes or peptides on an input protein sequence
|
|
Search for the HLA supertype-specific epitopes or binding peptides by protein sequence, the protein name could give an arbitrary name. The protein sequence should be a plain sequence. Such as: |
|
|
|
Search for mutated analogues of an input peptide
|
Search for the mutated analogues of an input peptide, amino acid sequence coding with uppercase letters. example : "FLPSDFFPSV". |
|
|
|
Search for promiscuous peptides presented by HLA molecules across supertypes
|
|
Search for promiscuous peptides to alleles across HLA supertypes by the supertyps or source species, |
|
|
|
NOTES
|
|
|
|
HLAsupE and HLAsupB
|
|
The query tools provided in the home page of HLAsupE aim to search for supertype-specific epitopes (T-cell activities), whereas the query tools for supertype-specific binding peptides (MHC-binding data) are provided in HLAsupB.
|
|
|
|
Epitopes with the restrictions of serological HLA molecules
|
|
The peptides presented by serological proteins were difficult to integrate into the supertype-specific dataset with an exact allele restriction. These data were only maintained in the T-cell activity and HLA-peptide binding data blocks; as a result, these peptide data can be displayed when a user inspects the detailed activity of a specific peptide, but a direct query of these peptides is currently unavailable in HLAsupE |
|
|
|
Frequency of alleles
|
The distribution of HLA genes in the human population varies with ethnicity and region. The frequency of alleles in a specific population should be taken into account. At present, Allele Frequency Net Database (AFND) and Population Coverage tool have been built to address this issue. |
|
|
|
Update
|
|
We will continue to update our database by extracting and curating HLA-restricted peptide data from all available epitope databases and published literature. We also encourage researchers to submit their own data to our database via email to wangshufeng81@hotmail.com. |
|
|
|
|
| 2018-2023©copyright Institute of Immunology, Army Medical University and Bioinformatics Center of Chongqing |
| contact: wangshufeng81@hotmail.com |